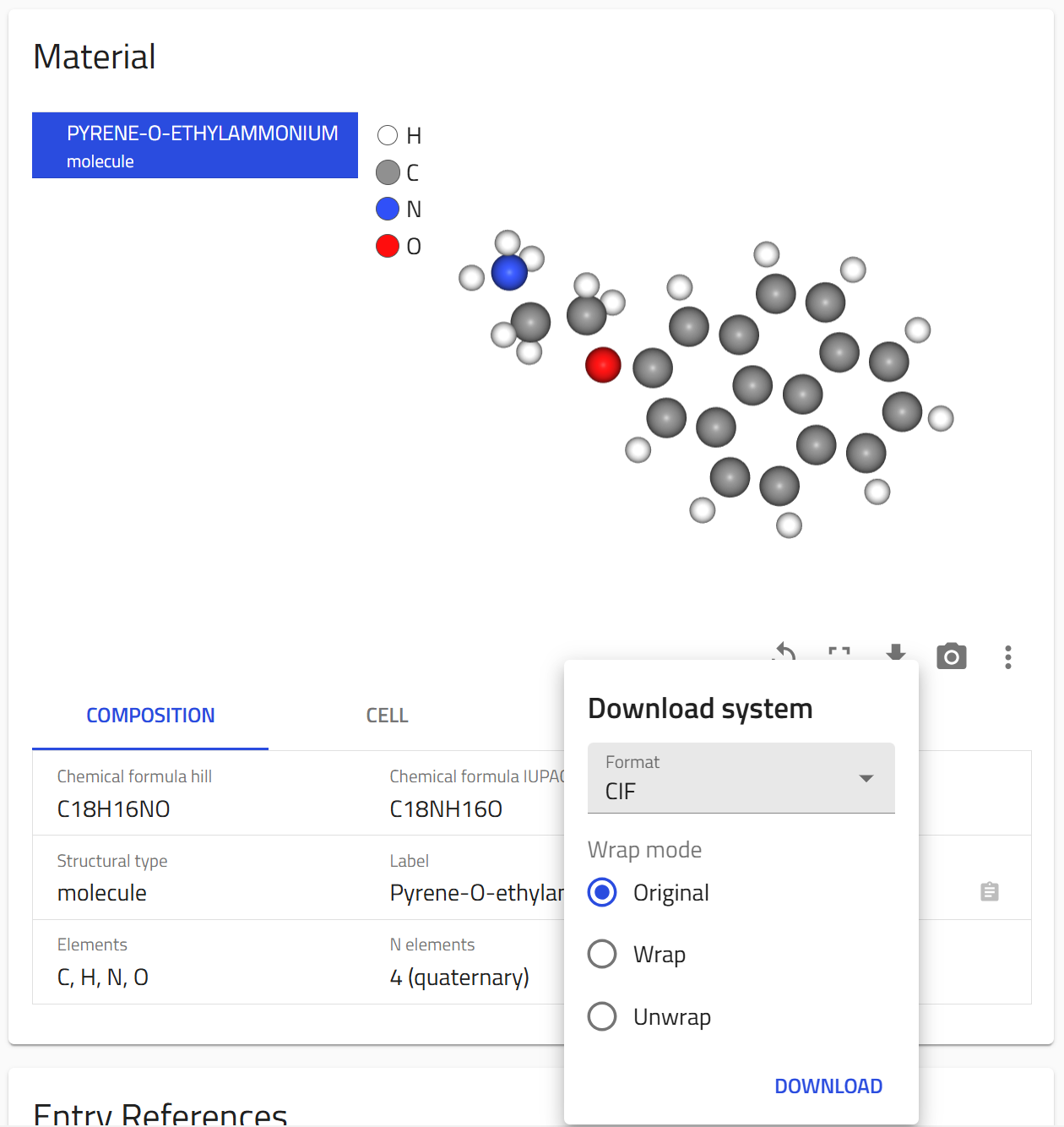
Exporting structure files¶
For the Halide Perovskite Ions Database we have calculated the conformers of the molecules using the RDKit Software. The structure files of these conformers can be downloaded directly from the overview page of the ion entry or Programmatically using the NOMAD API.
Downloading structure files from the overview page¶
Programmatically using the NOMAD API¶
Example for retrieving the structure of Pyrene-O-ethylammonium:
import requests
# Define the base URL
base_url = 'https://nomad-lab.eu/prod/v1/develop/api/v1/systems'
# Specify the entry ID
entry_id = 'ccun2zHQ49i6bA-RoTdK3U6mqBZM'
# Define the query parameters
params = {
'path': 'results/material/topology/0',
'format': 'xyz',
'wrap_mode': 'original'
}
# Construct the full URL by appending the entry ID
full_url = f'{base_url}/{entry_id}'
# Make the GET request with the specified parameters
response = requests.get(full_url, params=params)
# Check if the request was successful
if response.status_code == 200:
# Process the response content
data = response.content
# For example, save the content to a file
with open('output.xyz', 'wb') as file:
file.write(data)
else:
print(f'Error: {response.status_code}')
With this, you could write a script to download the structure files of the conformers of all the ions in the database.