Adding custom tasks to workflows using NOMAD's ELN Functionalities¶
Consider the following setup, simulation, and analysis protocol:
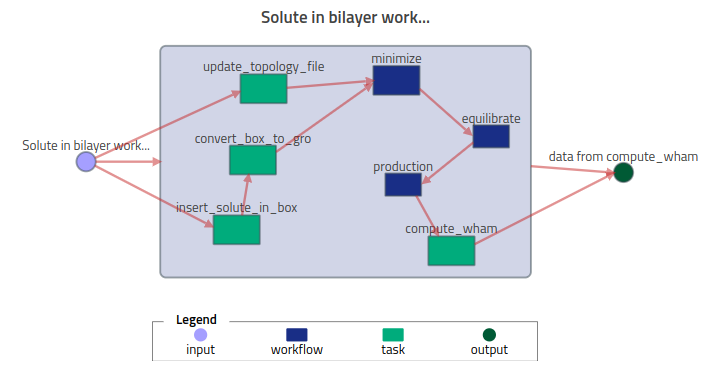
The minimize, equilibrate, and production (workflow) tasks are analogous to that described in Workflow - Simulation Protocol. The remaining tasks (green boxes) correspond to steps in the simulation protocol that are not supported by the NOMAD simulation parsers, e.g., creation of the initial configuration or model parameter files, or post-simulation analysis.
The basic strategy described here is to use NOMAD's existing ELN functionality to add (meta)data for these steps of the workflow. Here we only provide a description quantity, but more advanced support is available along this route.
Before we can add these as tasks in the workflow, we need to create mainfiles that NOMAD recognizes with the corresponding metadata. We can achieve this by creating an archive.yaml file as follows:
# insert_solute_in_box.archive.yaml
data:
m_def: nomad.datamodel.metainfo.eln.ElnBaseSection
name: 'insert_solute_in_box'
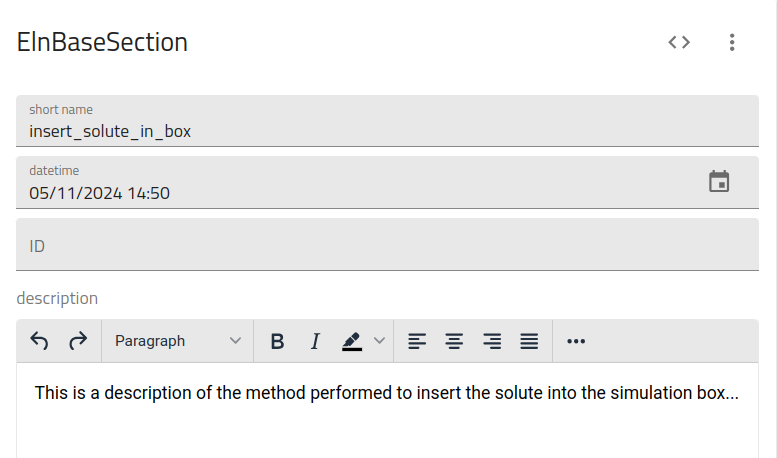
description: 'This is a description of the method performed to insert the solute into the simulation box...'
This utilizes the ELNBaseSection class to create the following overview page upon upload:
Now that we have a mainfile for each task, we can specify the graph strucuture and node attributes as described in the previous examples:
path_to_job = ''
node_attributes = {
0: {'name': 'Solute in bilayer workflow parameters',
'type': 'input',
'entry_type': 'other',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}workflow_parameters.archive.yaml',
},
'out_edge_nodes': [1, 3],
},
1: {'name': 'insert_solute_in_box',
'type': 'task',
'entry_type': 'other',
'path_info': {
'mainfile_path': f'{path_to_job}insert_solute_in_box.archive.yaml',
'archive_path': 'data',
},
'inputs': [
{
'name': 'data from workflow parameters',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}workflow_parameters.archive.yaml',
},
}
],
'outputs': [
{
'name': 'data from insert_solute_in_box',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}insert_solute_in_box.archive.yaml',
},
}
],
},
2: {'name': 'convert_box_to_gro',
'type': 'task',
'entry_type': 'other',
'path_info': {
'mainfile_path': f'{path_to_job}convert_box_to_gro.archive.yaml'
},
'in_edge_nodes': [1],
'inputs': [
{
'name': 'data from insert_solute_in_box',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}insert_solute_in_box.archive.yaml',
},
}
],
'outputs': [
{
'name': 'data from convert_box_to_gro',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}convert_box_to_gro.archive.yaml'
},
}
],
},
3: {'name': 'update_topology_file',
'type': 'task',
'entry_type': 'other',
'path_info': {
'mainfile_path': f'{path_to_job}update_topology_file.archive.yaml'
},
'inputs': [
{
'name': 'data from workflow parameters',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}workflow_parameters.archive.yaml',
},
}
],
'outputs': [
{
'name': 'data from update_topology_file',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}update_topology_file.archive.yaml'
},
}
],
},
4: {'name': 'minimize',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': f'{path_to_job}solute_in_bilayer_minimize.log'
},
'in_edge_nodes': [2, 3],
'inputs': [
{
'name': 'data from convert_box_to_gro',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}convert_box_to_gro.archive.yaml',
},
},
{
'name': 'data from update_topology_file',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}update_topology_file.archive.yaml',
},
}
],
},
5: {'name': 'equilibrate',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': f'{path_to_job}solute_in_bilayer_equilibrate.log'
},
'in_edge_nodes': [4],
},
6: {'name': 'production',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': f'{path_to_job}solute_in_bilayer_production.log'
},
'in_edge_nodes': [5],
},
7: {'name': 'compute_wham',
'type': 'task',
'entry_type': 'other',
'path_info': {
'mainfile_path': f'{path_to_job}compute_wham.archive.yaml'
},
'in_edge_nodes': [6],
'outputs': [
{
'name': 'data from compute_wham',
'path_info': {
'archive_path': 'data',
'mainfile_path': f'{path_to_job}compute_wham.archive.yaml'
},
}
],
},
}
and then apply the node_to_graph() and build_nomad_workflow() functions:
workflow_graph_input = nodes_to_graph(node_attributes)
workflow_metadata = {
'destination_filename': 'solute_in_bilayer.workflow.archive.yaml',
'workflow_name': 'Solute in bilayer workflow',
}
workflow_graph_output_minimal = build_nomad_workflow(
workflow_metadata=workflow_metadata,
workflow_graph=nx.DiGraph(workflow_graph_input_minimal),
write_to_yaml=True,
)
which produces the following workflow yaml:
'workflow2':
'name': 'Solute in bilayer workflow'
'inputs':
- 'name': 'Solute in bilayer workflow parameters'
'section': '../upload/archive/mainfile/workflow_parameters.archive.yaml#/data'
'outputs':
- 'name': 'data from compute_wham'
'section': '../upload/archive/mainfile/compute_wham.archive.yaml#/data'
'tasks':
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'insert_solute_in_box'
'task': '../upload/archive/mainfile/insert_solute_in_box.archive.yaml#/data'
'inputs':
- 'name': 'data from workflow parameters'
'section': '../upload/archive/mainfile/workflow_parameters.archive.yaml#/data'
'outputs':
- 'name': 'data from insert_solute_in_box'
'section': '../upload/archive/mainfile/insert_solute_in_box.archive.yaml#/data'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'convert_box_to_gro'
'inputs':
- 'name': 'data from insert_solute_in_box'
'section': '../upload/archive/mainfile/insert_solute_in_box.archive.yaml#/data'
'outputs':
- 'name': 'data from convert_box_to_gro'
'section': '../upload/archive/mainfile/convert_box_to_gro.archive.yaml#/data'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'update_topology_file'
'inputs':
- 'name': 'data from workflow parameters'
'section': '../upload/archive/mainfile/workflow_parameters.archive.yaml#/data'
'outputs':
- 'name': 'data from update_topology_file'
'section': '../upload/archive/mainfile/update_topology_file.archive.yaml#/data'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'minimize'
'task': '../upload/archive/mainfile/solute_in_bilayer_minimize.log#/workflow2'
'inputs':
- 'name': 'data from convert_box_to_gro'
'section': '../upload/archive/mainfile/convert_box_to_gro.archive.yaml#/data'
- 'name': 'input system from minimize'
'section': '../upload/archive/mainfile/solute_in_bilayer_minimize.log#/run/0/system/-1'
- 'name': 'data from update_topology_file'
'section': '../upload/archive/mainfile/update_topology_file.archive.yaml#/data'
'outputs':
- 'name': 'output system from minimize'
'section': '../upload/archive/mainfile/solute_in_bilayer_minimize.log#/run/0/system/-1'
- 'name': 'output calculation from minimize'
'section': '../upload/archive/mainfile/solute_in_bilayer_minimize.log#/run/0/calculation/-1'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'equilibrate'
'task': '../upload/archive/mainfile/solute_in_bilayer_equilibrate.log#/workflow2'
'inputs':
- 'name': 'input system from minimize'
'section': '../upload/archive/mainfile/solute_in_bilayer_minimize.log#/run/0/system/-1'
'outputs':
- 'name': 'output system from equilibrate'
'section': '../upload/archive/mainfile/solute_in_bilayer_equilibrate.log#/run/0/system/-1'
- 'name': 'output calculation from equilibrate'
'section': '../upload/archive/mainfile/solute_in_bilayer_equilibrate.log#/run/0/calculation/-1'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'production'
'task': '../upload/archive/mainfile/solute_in_bilayer_production.log#/workflow2'
'inputs':
- 'name': 'input system from equilibrate'
'section': '../upload/archive/mainfile/solute_in_bilayer_equilibrate.log#/run/0/system/-1'
'outputs':
- 'name': 'output system from production'
'section': '../upload/archive/mainfile/solute_in_bilayer_production.log#/run/0/system/-1'
- 'name': 'output calculation from production'
'section': '../upload/archive/mainfile/solute_in_bilayer_production.log#/run/0/calculation/-1'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'compute_wham'
'inputs':
- 'name': 'input system from production'
'section': '../upload/archive/mainfile/solute_in_bilayer_production.log#/run/0/system/-1'
'outputs':
- 'name': 'data from compute_wham'
'section': '../upload/archive/mainfile/compute_wham.archive.yaml#/data'