3 step molecular dynamics workflow for CG water¶
Consider the following 3 step MD equilibration procedure:
-
geometry optimization (energy minimization)
-
equilibration MD simulation in NPT
-
production MD simulation in NVT
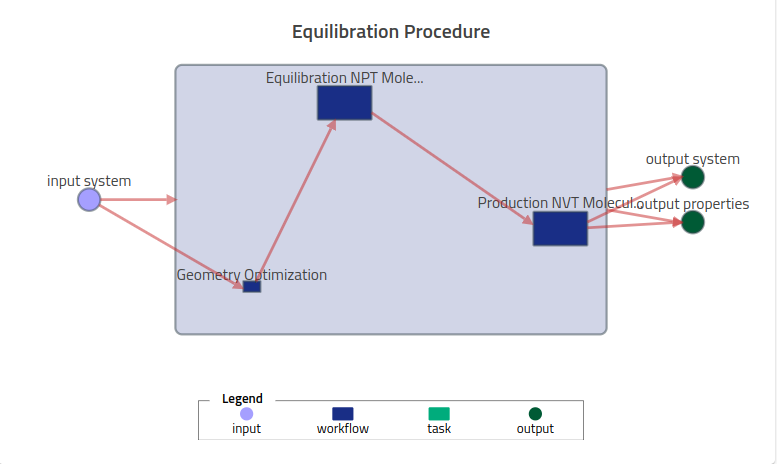
Each task in this workflow represents a supported entry in NOMAD, and all 3 simulations will be uploaded together with the workflow.archive.yaml file, within the following local filesystem:
upload.zip
├── workflow.archive.yaml
├── Emin
│ ├── mdrun_Emin.log # Geometry Optimization mainfile
│ └── ...other raw simulation files
├── Equil_NPT
│ ├── mdrun_Equil-NPT.log # NPT equilibration mainfile
│ └── ...other raw simulation files
└── Prod_NVT
├── mdrun_Prod-NVT.log # NVT production mainfile
└── ...other raw simulation files
The following will demonstrate how to generate the yaml file automatically with nomad-utility-workflows.
First import the necessary imports (gravis is only used for graph visualization and is not strictly necessary):
import gravis as gv
import networkx as nx
from nomad_utility_workflows.utils.workflows import build_nomad_workflow, nodes_to_graph
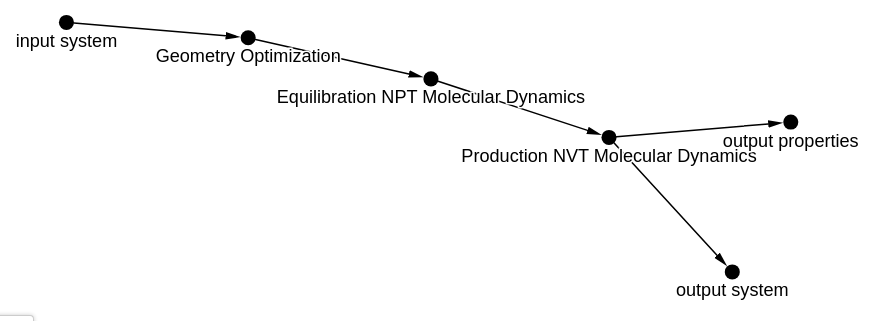
To generate the appropriate workflow yaml file to connect the entries in NOMAD, we need to create a graph representating
our workflow. For this workflow, we create a networkx.DiGraph()
which we name workflow_graph_input that looks like:
IMPORTANT
To ensure that all of our functionalities work correctly, the node keys should simply be unique integers that index the nodes. I.e., node_keys = [0, 1, 2, 3] for a graph with 4 nodes.
If you are using a workflow manager, you should be able to extract a graph structure directly from the manager output, and then map this structure to an analogous structure with networkx, as described further below. If you do not have access to such a graph, you can generate the nx.DiGraph manually (see ...) or alternatively use our nodes_to_graph() functionality described in Below.
A series of attributes should be associated with each of the nodes in your graph. Here we print out these node attributes from the graph visualized above:
for node_key, node_attributes in workflow_graph_input.nodes(data=True):
print(node_key, node_attributes)
0 {'name': 'input system',
'type': 'input',
'path_info': {
'mainfile_path': 'Emin/mdrun_Emin.log',
'supersection_index': 0,
'section_index': 0,
'section_type': 'system'
},
}
1 {'name': 'Geometry Optimization',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Emin/mdrun_Emin.log'
}
}
2 {'name': 'Equilibration NPT Molecular Dynamics',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'
},
}
3 {'name': 'Production NVT Molecular Dynamics',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
}
4 {'name': 'output system',
'type': 'output',
'path_info': {
'section_type': 'system',
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
}
5 {'name': 'output properties',
'type': 'output',
'path_info': {
'section_type': 'calculation',
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
}
Descriptions for each attribute are given in Explanation > Workflow
The appropriate edges should be added to your graph:
for edge_1, edge_2, edge_attributes in workflow_graph_input.edges(data=True):
print(edge_1, edge_2, edge_attributes)
There are no necessary attributes to add to the graph. nomad-utility-workflows will automatically add edge attributes based on the node attribute inputs.
Now that we have generated the input workflow graph, we can use nomad-utility-workflows's build_nomad_workflow() function to create the workflow.archive.yaml file that will connect our individual simulations within NOMAD:
workflow_metadata = {
'destination_filename': './workflow_minimal.archive.yaml',
'workflow_name': 'Equilibration Procedure',
}
workflow_graph_output = build_nomad_workflow(
workflow_metadata=workflow_metadata,
workflow_graph=nx.DiGraph(workflow_graph_input),
write_to_yaml=True,
)
Here we provide the full path and name of the output yaml in destination_filename and the overarching workflow name that will show up on top of the workflow graph visualization in workflow_name. The output workflow looks like:
gv.d3(
workflow_graph_output,
node_label_data_source='name',
edge_label_data_source='name',
zoom_factor=1.5,
node_hover_tooltip=True,
)
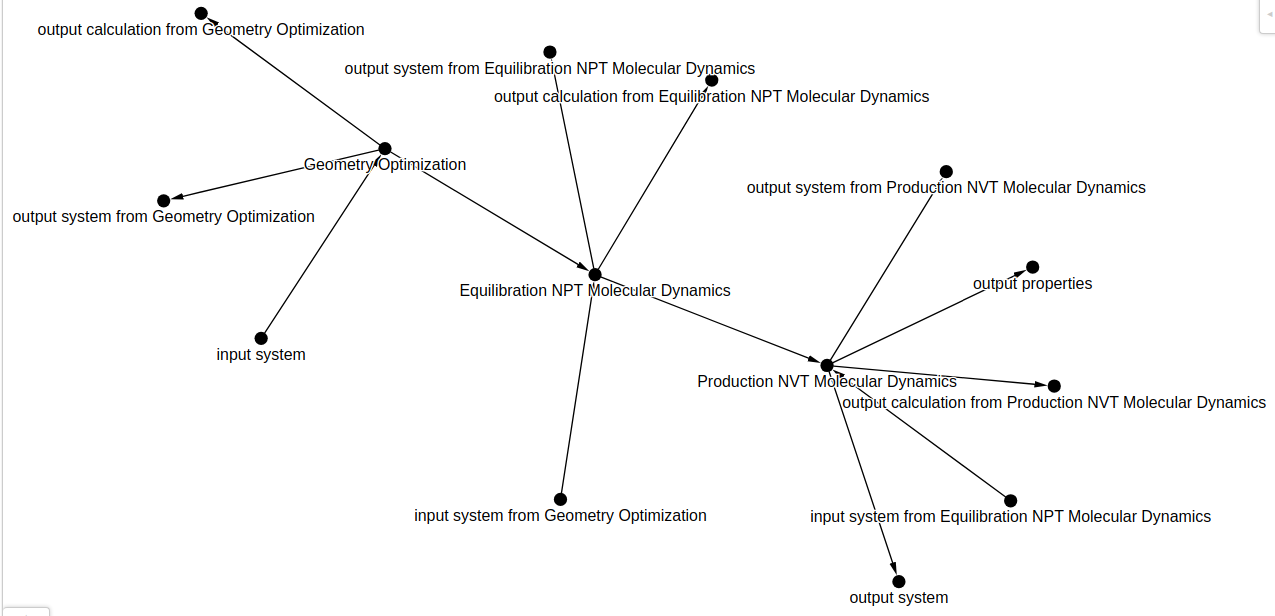
We see that our output graph looks signficantly different than the input. That's because nomad-utility-workflow is automatically adding some default input/outputs to ensure the proper node connections within the workflow visualizer. For simulations, the automatically generated input defaults correspond to the system section from any incoming task node that exists. The automatically generated output defaults correspond to both the system and the calculation section from the given node.
Let's examine the output workflow graph in more detail:
for node_key, node_attributes in workflow_graph_output_minimal.nodes(data=True):
print(node_key, node_attributes)
0 {'name': 'input system', 'type': 'input', 'path_info': {'mainfile_path': 'Emin/mdrun_Emin.log', 'supersection_index': 0, 'section_index': 0, 'section_type': 'system'}}
1 {'name': 'Geometry Optimization', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Emin/mdrun_Emin.log'}}
2 {'name': 'Equilibration NPT Molecular Dynamics', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
3 {'name': 'Production NVT Molecular Dynamics', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
4 {'name': 'output system', 'type': 'output', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
5 {'name': 'output properties', 'type': 'output', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
6 {'type': 'output', 'name': 'output system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}
7 {'type': 'output', 'name': 'output calculation from Geometry Optimization', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Emin/mdrun_Emin.log'}}
8 {'type': 'input', 'name': 'input system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}
9 {'type': 'output', 'name': 'output system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
10 {'type': 'output', 'name': 'output calculation from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
11 {'type': 'input', 'name': 'input system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
12 {'type': 'output', 'name': 'output system from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
13 {'type': 'output', 'name': 'output calculation from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
and the edges:
for edge_1, edge_2, edge_attributes in workflow_graph_output_minimal.edges(data=True):
print(edge_1, edge_2, edge_attributes)
0 1 {'inputs': [], 'outputs': []}
1 2 {'inputs': [{'name': 'output system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}, {'name': 'output calculation from Geometry Optimization', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Emin/mdrun_Emin.log'}}], 'outputs': [{'name': 'input system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}]}
1 6 {}
1 7 {}
2 3 {'inputs': [{'name': 'output system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}, {'name': 'output calculation from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}], 'outputs': [{'name': 'input system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}]}
2 9 {}
2 10 {}
3 4 {'inputs': [{'name': 'output system from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}, {'name': 'output calculation from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}], 'outputs': []}
3 5 {'inputs': [], 'outputs': []}
3 12 {}
3 13 {}
8 2 {}
11 3 {}
Finally, the output workflow.archive.yaml:
'workflow2':
'name': 'Equilibration Procedure'
'inputs':
- 'name': 'input system'
'section': '../upload/archive/mainfile/Emin/mdrun_Emin.log#/run/0/system/0'
'outputs':
- 'name': 'output system'
'section': '../upload/archive/mainfile/Prod_NVT/mdrun_Prod-NVT.log#/run/0/system/-1'
- 'name': 'output properties'
'section': '../upload/archive/mainfile/Prod_NVT/mdrun_Prod-NVT.log#/run/0/calculation/-1'
'tasks':
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'Geometry Optimization'
'task': '../upload/archive/mainfile/Emin/mdrun_Emin.log#/workflow2'
'inputs': []
'outputs':
- 'name': 'output system from Geometry Optimization'
'section': '../upload/archive/mainfile/Emin/mdrun_Emin.log#/run/0/system/-1'
- 'name': 'output calculation from Geometry Optimization'
'section': '../upload/archive/mainfile/Emin/mdrun_Emin.log#/run/0/calculation/-1'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'Equilibration NPT Molecular Dynamics'
'task': '../upload/archive/mainfile/Equil_NPT/mdrun_Equil-NPT.log#/workflow2'
'inputs':
- 'name': 'input system from Geometry Optimization'
'section': '../upload/archive/mainfile/Emin/mdrun_Emin.log#/run/0/system/-1'
'outputs':
- 'name': 'output system from Equilibration NPT Molecular Dynamics'
'section': '../upload/archive/mainfile/Equil_NPT/mdrun_Equil-NPT.log#/run/0/system/-1'
- 'name': 'output calculation from Equilibration NPT Molecular Dynamics'
'section': '../upload/archive/mainfile/Equil_NPT/mdrun_Equil-NPT.log#/run/0/calculation/-1'
- 'm_def': 'nomad.datamodel.metainfo.workflow.TaskReference'
'name': 'Production NVT Molecular Dynamics'
'task': '../upload/archive/mainfile/Prod_NVT/mdrun_Prod-NVT.log#/workflow2'
'inputs':
- 'name': 'input system from Equilibration NPT Molecular Dynamics'
'section': '../upload/archive/mainfile/Equil_NPT/mdrun_Equil-NPT.log#/run/0/system/-1'
'outputs':
- 'name': 'output system from Production NVT Molecular Dynamics'
'section': '../upload/archive/mainfile/Prod_NVT/mdrun_Prod-NVT.log#/run/0/system/-1'
- 'name': 'output calculation from Production NVT Molecular Dynamics'
'section': '../upload/archive/mainfile/Prod_NVT/mdrun_Prod-NVT.log#/run/0/calculation/-1'
and the resulting workflow visualization in NOMAD:
Creating an input graph with nodes_to_graph()¶
If you are unfamiliar with networkx, nomad-utility-workflows can generate an input graph structure for you to use with build_workflow_yaml().
For this approach, simply create a dictionary of node keys and corresponding attributes:
node_attributes = {
0: {'name': 'input system',
'type': 'input',
'path_info': {
'mainfile_path': 'Emin/mdrun_Emin.log',
'supersection_index': 0,
'section_index': 0,
'section_type': 'system'
},
'out_edge_nodes': [1],
},
1: {'name': 'Geometry Optimization',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Emin/mdrun_Emin.log'
}
},
2: {'name': 'Equilibration NPT Molecular Dynamics',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'
},
'in_edge_nodes': [1],
},
3: {'name': 'Production NVT Molecular Dynamics',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
'in_edge_nodes': [2],
},
4: {'name': 'output system',
'type': 'output',
'path_info': {
'section_type': 'system',
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
'in_edge_nodes': [3],
},
5: {'name': 'output properties',
'type': 'output',
'path_info': {
'section_type': 'calculation',
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
'in_edge_nodes': [3],
}
}
Notice that this dictionary exactly corresponds to the printed attributes of the nodes displayed above, with the exception of the in_edge_nodes and out_edge_nodes attributes. These attributes are used to specify the graph edges.
Now, simply run:
The resulting graph should be identical to the input graph visualized above, and can be therefore used in the same way to generate the workflow yaml file.
Adding additional inputs/outputs¶
Now let's add some additional input/outputs to the nodes. We will use the nodes_to_graph() method to generate the input workflow graph:
node_attributes = {
0: {'name': 'input system',
'type': 'input',
'path_info': {
'mainfile_path': 'Emin/mdrun_Emin.log',
'supersection_index': 0,
'section_index': 0,
'section_type': 'system'
},
'out_edge_nodes': [1],
},
1: {'name': 'Geometry Optimization',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Emin/mdrun_Emin.log'
},
'outputs': [
{
'name': 'energies of the relaxed system',
'path_info': {
'section_type': 'energy',
'supersection_path': 'run/0/calculation', # this can be done, but at this point it's safer / easier to just use archive_path
'supersection_index': -1,
},
}
],
},
2: {'name': 'Equilibration NPT Molecular Dynamics',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'
},
'in_edge_nodes': [1],
'outputs': [
{
'name': 'MD workflow properties (structural and dynamical)',
'path_info': {
'section_type': 'results',
},
}
],
},
3: {'name': 'Production NVT Molecular Dynamics',
'type': 'workflow',
'entry_type': 'simulation',
'path_info': {
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
'in_edge_nodes': [2],
'outputs': [
{
'name': 'MD workflow properties (structural and dynamical)',
'path_info': {
'section_type': 'results',
},
}
],
},
4: {'name': 'output system',
'type': 'output',
'path_info': {
'section_type': 'system',
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
'in_edge_nodes': [3],
},
5: {'name': 'output properties',
'type': 'output',
'path_info': {
'section_type': 'calculation',
'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'
},
'in_edge_nodes': [3],
}
}
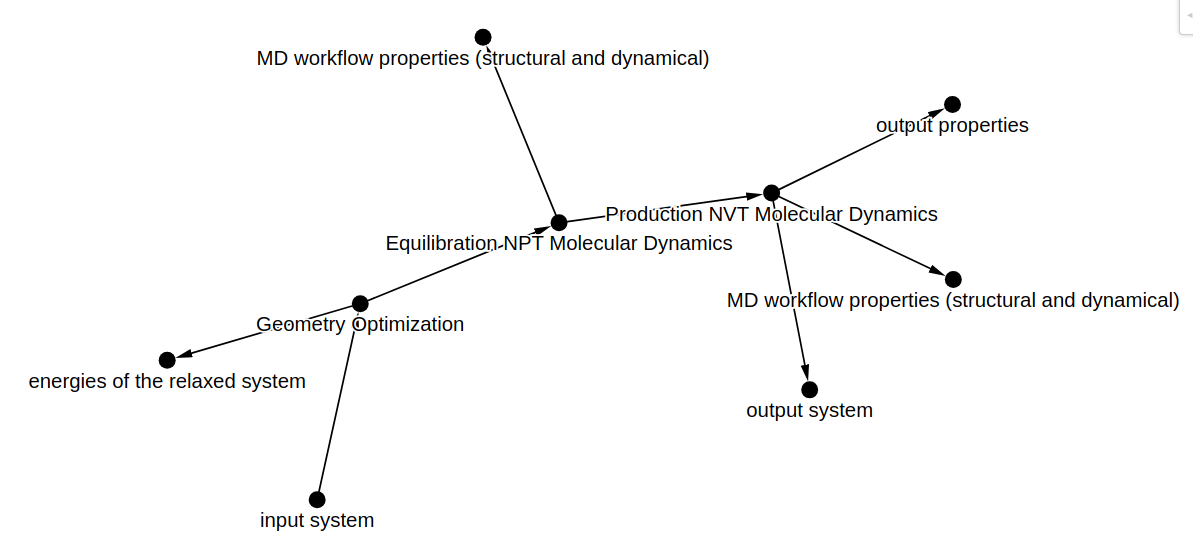
workflow_graph_input = nodes_to_graph(node_attributes)
gv.d3(
workflow_graph_input,
node_label_data_source='name',
edge_label_data_source='name',
zoom_factor=1.5,
node_hover_tooltip=True,
)
for node_key, node_attributes in workflow_graph_input.nodes(data=True):
print(node_key, node_attributes)
0 {'name': 'input system', 'type': 'input', 'path_info': {'mainfile_path': 'Emin/mdrun_Emin.log', 'supersection_index': 0, 'section_index': 0, 'section_type': 'system'}, 'out_edge_nodes': [1]}
1 {'name': 'Geometry Optimization', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Emin/mdrun_Emin.log'}}
2 {'name': 'Equilibration NPT Molecular Dynamics', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}, 'in_edge_nodes': [1]}
3 {'name': 'Production NVT Molecular Dynamics', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}, 'in_edge_nodes': [2]}
4 {'name': 'output system', 'type': 'output', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}, 'in_edge_nodes': [3]}
5 {'name': 'output properties', 'type': 'output', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}, 'in_edge_nodes': [3]}
6 {'type': 'output', 'name': 'energies of the relaxed system', 'path_info': {'section_type': 'energy', 'supersection_path': 'run/0/calculation', 'supersection_index': -1}}
7 {'type': 'output', 'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results'}}
8 {'type': 'output', 'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results'}}
for edge_1, edge_2, edge_attributes in workflow_graph_input.edges(data=True):
print(edge_1, edge_2, edge_attributes)
0 1 {}
1 6 {'inputs': [{'name': 'energies of the relaxed system', 'path_info': {'section_type': 'energy', 'supersection_path': 'run/0/calculation', 'supersection_index': -1}}]}
1 2 {}
2 7 {'inputs': [{'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results'}}]}
2 3 {}
3 8 {'inputs': [{'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results'}}]}
3 4 {}
3 5 {}
And again we generate the final workflow graph and yaml:
workflow_graph_output = build_nomad_workflow(
destination_filename='./workflow_minimal.archive.yaml',
workflow_name='Equilibration Procedure',
workflow_graph=nx.DiGraph(workflow_graph_input),
write_to_yaml=True,
)
gv.d3(
workflow_graph_output,
node_label_data_source='name',
edge_label_data_source='name',
zoom_factor=1.5,
node_hover_tooltip=True,
)
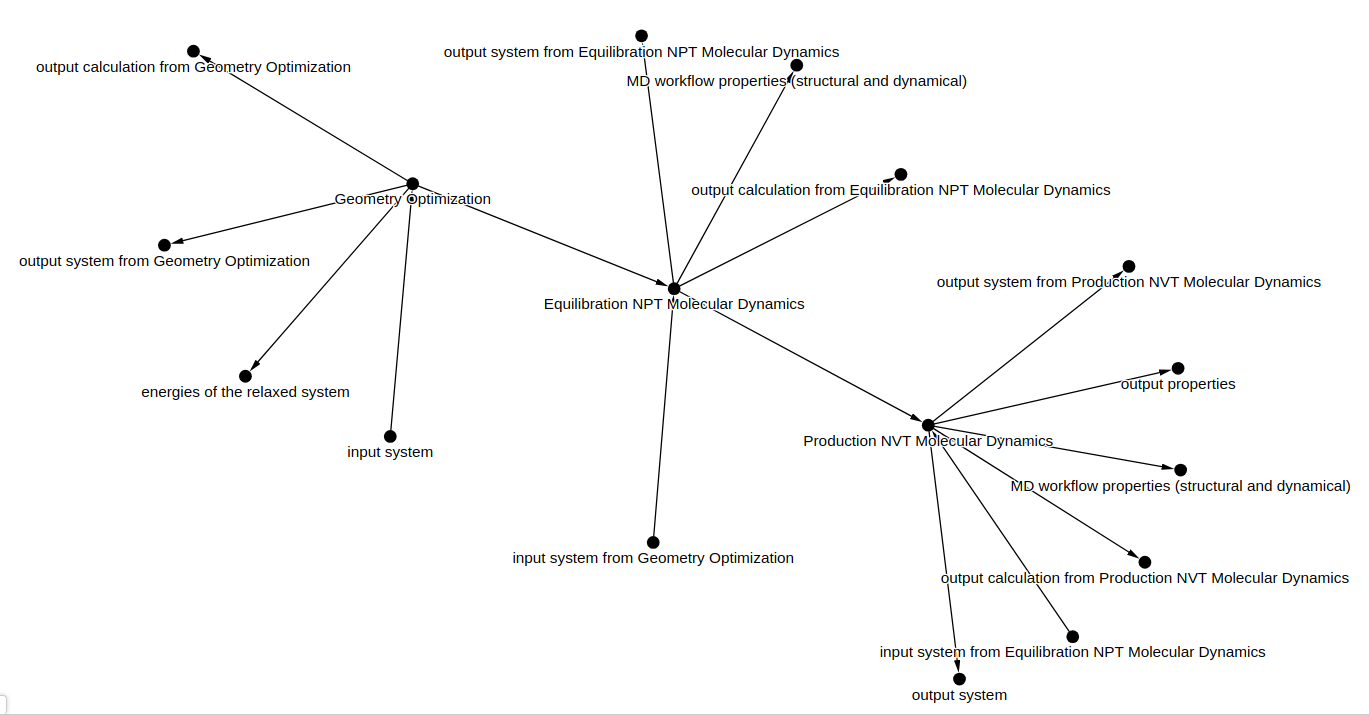
for node_key, node_attributes in workflow_graph_output.nodes(data=True):
print(node_key, node_attributes)
0 {'name': 'input system', 'type': 'input', 'path_info': {'mainfile_path': 'Emin/mdrun_Emin.log', 'supersection_index': 0, 'section_index': 0, 'section_type': 'system'}, 'out_edge_nodes': [1]}
1 {'name': 'Geometry Optimization', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Emin/mdrun_Emin.log'}}
2 {'name': 'Equilibration NPT Molecular Dynamics', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}, 'in_edge_nodes': [1]}
3 {'name': 'Production NVT Molecular Dynamics', 'type': 'workflow', 'entry_type': 'simulation', 'path_info': {'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}, 'in_edge_nodes': [2]}
4 {'name': 'output system', 'type': 'output', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}, 'in_edge_nodes': [3]}
5 {'name': 'output properties', 'type': 'output', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}, 'in_edge_nodes': [3]}
6 {'type': 'output', 'name': 'energies of the relaxed system', 'path_info': {'section_type': 'energy', 'supersection_path': 'run/0/calculation', 'supersection_index': -1, 'mainfile_path': 'Emin/mdrun_Emin.log'}}
7 {'type': 'output', 'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
8 {'type': 'output', 'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
9 {'type': 'output', 'name': 'output system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}
10 {'type': 'output', 'name': 'output calculation from Geometry Optimization', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Emin/mdrun_Emin.log'}}
11 {'type': 'input', 'name': 'input system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}
12 {'type': 'output', 'name': 'output system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
13 {'type': 'output', 'name': 'output calculation from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
14 {'type': 'input', 'name': 'input system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}
15 {'type': 'output', 'name': 'output system from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
16 {'type': 'output', 'name': 'output calculation from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}
for edge_1, edge_2, edge_attributes in workflow_graph_output.edges(data=True):
print(edge_1, edge_2, edge_attributes)
0 1 {'inputs': [], 'outputs': []}
1 6 {'inputs': [{'name': 'energies of the relaxed system', 'path_info': {'section_type': 'energy', 'supersection_path': 'run/0/calculation', 'supersection_index': -1, 'mainfile_path': 'Emin/mdrun_Emin.log'}}, {'name': 'output system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}, {'name': 'output calculation from Geometry Optimization', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Emin/mdrun_Emin.log'}}], 'outputs': []}
1 2 {'inputs': [], 'outputs': [{'name': 'input system from Geometry Optimization', 'path_info': {'section_type': 'system', 'mainfile_path': 'Emin/mdrun_Emin.log'}}]}
1 9 {}
1 10 {}
2 7 {'inputs': [{'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}, {'name': 'output system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}, {'name': 'output calculation from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}], 'outputs': []}
2 3 {'inputs': [], 'outputs': [{'name': 'input system from Equilibration NPT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Equil_NPT/mdrun_Equil-NPT.log'}}]}
2 12 {}
2 13 {}
3 8 {'inputs': [{'name': 'MD workflow properties (structural and dynamical)', 'path_info': {'section_type': 'results', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}, {'name': 'output system from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'system', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}, {'name': 'output calculation from Production NVT Molecular Dynamics', 'path_info': {'section_type': 'calculation', 'mainfile_path': 'Prod_NVT/mdrun_Prod-NVT.log'}}], 'outputs': []}
3 4 {'inputs': [], 'outputs': []}
3 5 {'inputs': [], 'outputs': []}
3 15 {}
3 16 {}
11 2 {}
14 3 {}