How to Use This Plugin¶
This plugin can be used in a NOMAD Oasis installation. Check the tips in Install this plugin for details.
Once the NOMAD Oasis is deployed, you can generate different entries as shown below.
Outline¶
- Create an upload and manage users
- Create a new sample entry
- Create a sample processing conditions entry
- Add an instrument entry
- Add a measurement entry
- Upload a DFT calculation
- Create an ExperimentELN entry
- Use the dedicated UniSysCat Explore App
- Run a jupyter notebook to visualize the
.pdbfile
1. Create an upload and manage users¶
2. Create a new sample entry¶
3. Create sample processing conditions¶
4. Add an instrument entry¶
5. Add a measurement entry¶
6. Upload a DFT calculation¶
7. Create an ExperimentELN entry¶
You can create an Experiment entry and link all related activities in one entry, similar to the sample processing entry. Select ExperimentELN Entry from the drop-down menu and link the references in the subsection "steps"
8. Use the dedicated UniSysCat Explore App to get an overview and find entries¶
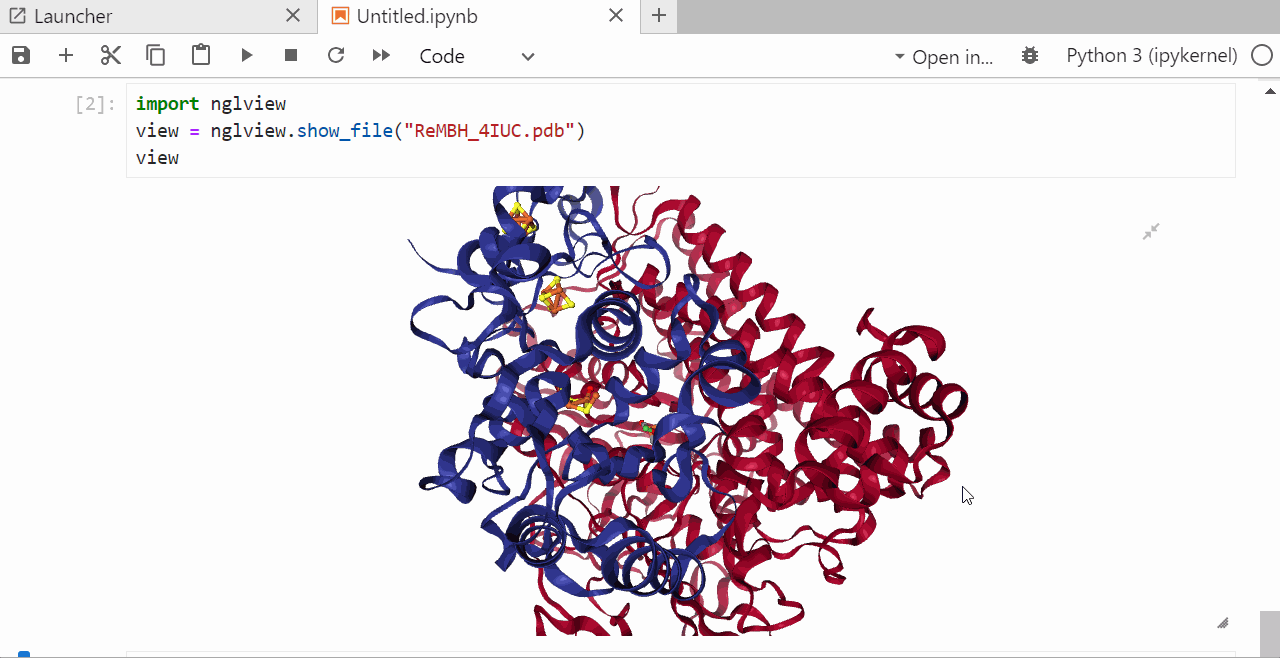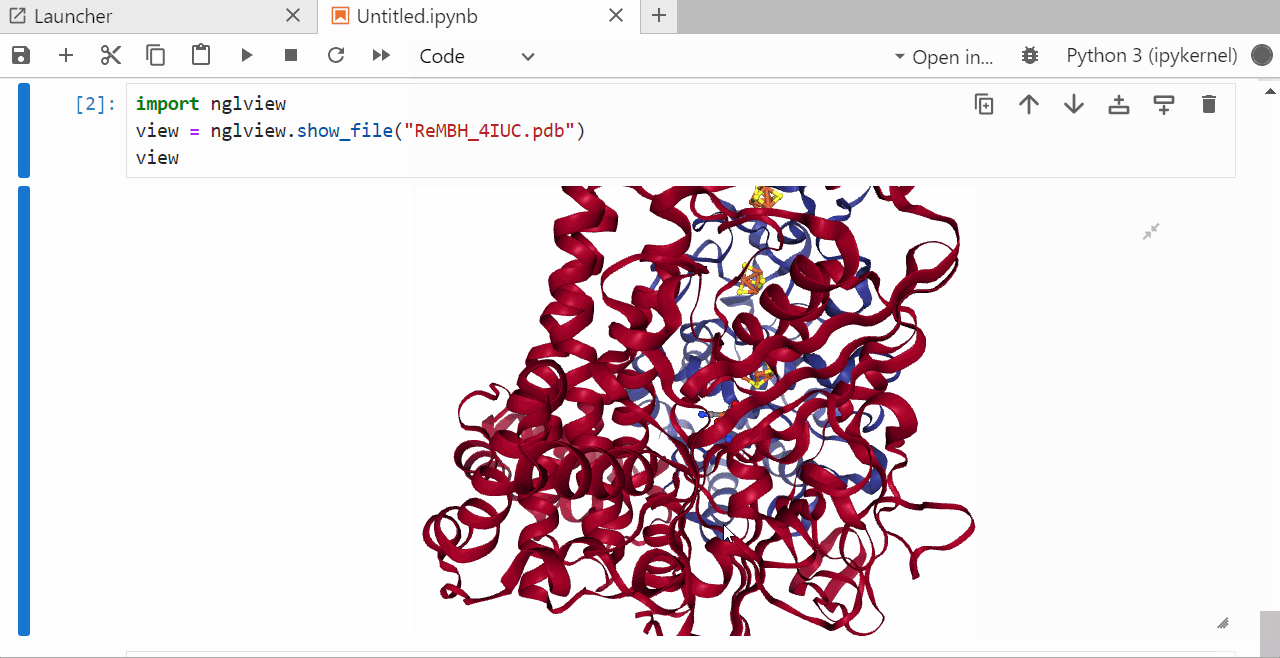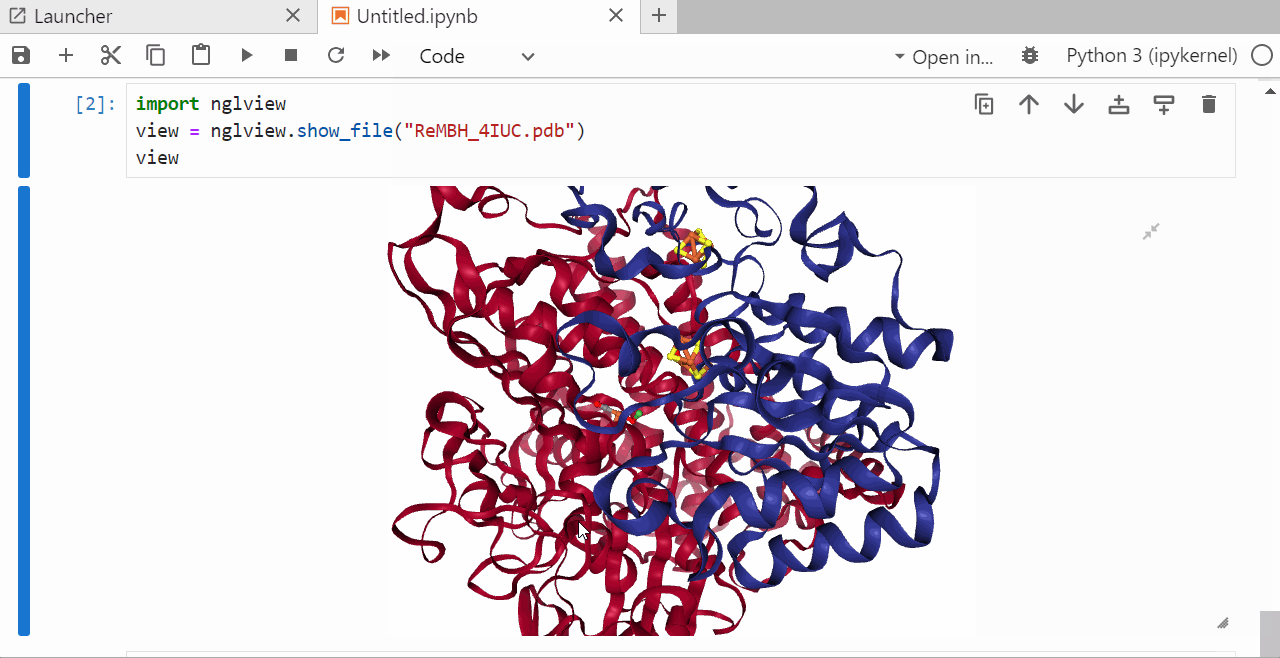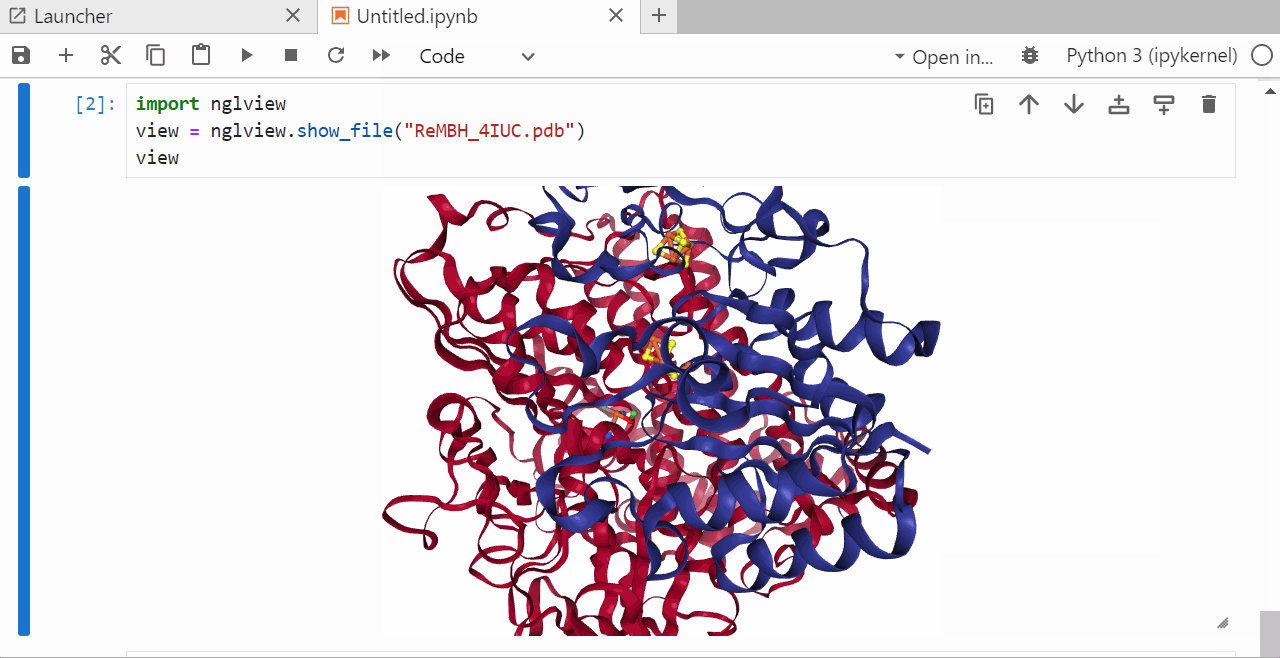
9. Run a jupyter notebook to visualize the .pdb file¶
In the open jupyter notebook install the ngl package by typing
pip install nglview shift +enter